Manufacturing and Analytical Characterization - Biomolecular
(M1230-02-09) Separation and Structural Analysis of Heavily Modified Therapeutic RNAs Using Hydrophilic Interaction Liquid Chromatography Coupled to Advanced Ion Activation Techniques for Tandem Mass Spectrometry
Monday, November 10, 2025
12:30 PM - 1:30 PM CT
- MG
Mohamed I. I. Gadallah, MS (he/him/his)
PhD Graduate Student
University of Texas at Austin
Austin, Texas, United States - MG
Mohamed I. I. Gadallah, MS (he/him/his)
PhD Graduate Student
University of Texas at Austin
Austin, Texas, United States - NE
Noha M. El Zahar, PhD
Associate porfessor
Ain Shams University
Cairo, Al Qahirah, Egypt - TN
Thanh Nguyen, BS
PhD Graduate Student
University of Texas at Austin
Austin, Texas, United States - HR
Hanlin Ren, BS
PhD Graduate Student
University of Texas at Austin
Austin, Texas, United States - JB
Jennifer S. Brodbelt, Ph.D.
Department Chair
University of Texas at Austin
Austin, Texas, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: Therapeutic RNA represents a rapidly advancing modality capable of targeting diseases that were previously considered undruggable by conventional small molecules.1,2 RNAs biological efficacy is critically dependent on the targeted delivery of the intact molecules to the site of action. To achieve this, therapeutic RNAs are heavily chemically modified — for example, by incorporating 2'-O-methyl and 2'-fluoro modifications and/or phosphorothioate backbones to enhance nuclease resistance 3,4— and often conjugated with targeting moieties such as N-acetylgalactosamine to enable receptor-mediated uptake by hepatocytes.5,6 Therefore, accurate characterization of these chemical modifications is essential to ensure integrity, stability, and correct functionalization of the therapeutic agent. Currently, ion-pairing reversed-phase liquid chromatography (IP-RPLC) coupled with mass spectrometry is the predominant method for RNA analysis.7 However, this approach suffers from some drawbacks, including ion suppression and contamination of the mass spectrometer due to the use of non-volatile ion-pairing reagents.8 Hydrophilic interaction liquid chromatography (HILIC) has emerged as a promising alternative, offering enhanced MS compatibility, reduced ion suppression, and improved sensitivity for the analysis of therapeutic RNAs.9,10 In this study, we developed and implemented a HILIC-MS method coupled with different collisional and photon-based activation techniques to characterize heavily chemically-modified RNA therapeutics with high sensitivity and structural fidelity.
Methods: Chemically-modified Patisiran RNA strands were obtained from Integrated DNA Technologies (Coralville, IA), ATU027 and Fitusiran siRNAs were purchased from MedChemExpress. Samples were diluted to a final concentration of 10 µM in 10 mM piperidine and used without further purification. HILIC separation of therapeutic RNA was performed using a GTxResolve Premier BEH Amide column (300 Å, 1.7 µm, 2.1 × 150 mm; Waters) on a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific) using 25% acetonitrile (MPA) and 95% LC-MS acetonitrile (MPB) both with 10 mM ammonium acetate. MS analysis was performed using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) source operating in negative ion mode. The mass spectrometer was modified with a 193 nm excimer laser (Coherent Inc., 500 Hz) to enable ultraviolet photodissociation (UVPD) for MS/MS. For structural characterization, collision induced dissociation (CID) was performed using NCE ranging from 15 to 20. UVPD was conducted using a single laser pulse at 2-3 mJ to generate diverse fragment ions, while activated-electron photodetachment (a-EPD) was achieved with a single 1 mJ laser pulse followed by supplemental collisional activation (NCE 10–15). Mass spectral data were processed and interpreted using the Nucleo-SAFARI software platform.11
Results: A HILIC-MS workflow was established for the analysis of modified therapeutic RNAs, enabling effective separation of the sense and antisense strands of Patisiran without reliance on non-volatile ion-pairing reagents (Figure 1). The use of 10 mM ammonium acetate as a volatile mobile phase additive provided excellent chromatographic performance, yielding sharp, well-resolved peaks and minimizing salt adduction—thereby enhancing compatibility with mass spectrometric detection. CID of single strands achieved complete (100%) sequence coverage for both sense and antisense RNA, with fragmentation dominated by c- and y-type ions and a large amount of base-loss products (Figure 2). This indicates highly efficient backbone cleavage and confirms that CID is well suited for routine characterization of canonical phosphodiester backbones. In contrast, UVPD and EPD produced a more diverse array of fragment ions—including a-, w-, and d-type species as well as c- and y- fragments—which provided broader structural coverage across the RNA molecule. These techniques enabled the mapping of modifications located along the phosphate backbone, ribose sugar, and nitrogenous bases, offering a more comprehensive representation of RNA architecture than CID alone. When analyzing double-stranded siRNA, the HILIC method successfully preserved the native duplex, observed as a distinct peak in the chromatogram. Gentle in-source activation disrupted the duplex with minimum fragmentation, and quadrupole isolation using narrow m/z windows enabled strand-specific isolation prior to MS/MS (Figure 3). Under these conditions, CID provided 75% and 87% sequence coverage for the sense and antisense strands, respectively. However, the combination of CID and UVPD increased sequence coverage to 91% for the sense and 95% for the antisense strands—surpassing previously reported values for similar sizes of RNAs and highlighting the advantages of multimodal activation.12 Collectively, these results demonstrate that coupling HILIC-MS with complementary gas-phase activation techniques enables structural characterization of therapeutic RNA. This integrated approach offers a significant improvement over conventional IP-RPLC–MS workflows and is well suited for both discovery-stage and regulatory applications in RNA therapeutic development.
Conclusion: HILIC separation enabled effective resolution of intact therapeutic RNA strands from low-abundance degradation products without the need for non-volatile ion-pairing reagents, ensuring full compatibility with mass spectrometry detection. Coupling this chromatographic platform with advanced ion activation techniques provided comprehensive structural characterization, supporting accurate sequencing and validation of chemical modifications critical for RNA therapeutic development and quality assessment.
References: (1) Androsavich, J. R. Frameworks for Transformational Breakthroughs in RNA-Based Medicines. Nature Reviews Drug Discovery 2024 23:6 2024, 23 (6), 421–444.
(2) Saw, P. E.; Song, E. Advancements in Clinical RNA Therapeutics: Present Developments and Prospective Outlooks. Cell Rep Med 2024, 5 (5), 101555.
(3) Shi, Y.; Zhen, X.; Zhang, Y.; Li, Y.; Koo, S.; Saiding, Q.; Kong, N.; Liu, G.; Chen, W.; Tao, W. Chemically Modified Platforms for Better RNA Therapeutics. Chem Rev 2024, 124 (3), 929–1033.
(4) Delaunay, S.; Helm, M.; Frye, M. RNA Modifications in Physiology and Disease: Towards Clinical Applications. Nature Reviews Genetics 2023 25:2 2023, 25 (2), 104–122.
(5) Li, Q.; Yin, K.; Ma, H. P.; Liu, H. H.; Li, S.; Luo, X.; Hu, R.; Zhang, W. W.; Lv, Z. S.; Niu, X. L.; Gu, M. H.; Li, C. L.; Liu, Y. S.; Liu, Y. J.; Li, H. B.; Li, N.; Li, C.; Gu, W. W.; Li, J. J. Application of Improved GalNAc Conjugation in Development of Cost-Effective SiRNA Therapies Targeting Cardiovascular Diseases. Molecular Therapy 2024, 32 (3), 637–645.
(6) Kashyap, D.; Booth, M. J. Nucleic Acid Conjugates: Unlocking Therapeutic Potential. ACS Bio and Med Chem Au 2024.
(7) Maurer, J.; Malburet, C.; François-Heude, M.; Guillarme, D. Evaluation of Ion Pairing Reversed-Phase Liquid Chromatography for the Separation of Large RNA Molecules. J Chromatogr A 2025, 1740, 465574.
(8) Hayashi, Y.; Sun, Y. Overcoming Challenges in Oligonucleotide Therapeutics Analysis: A Novel Nonion Pair Approach. J Am Soc Mass Spectrom 2024, 35 (9), 2034–2037.
(9) Hsiao, J. J.; Bertram, L. J.; Apffel, A.; Tripodi, A. A. P.; Coffey, A.; Wei, T.-C.; Flannery, C. Systematic Evaluation of HILIC Stationary Phases for MS Characterization of Oligonucleotides. LCGC International 2024, 10–20.
(10) Lardeux, H.; D’Atri, V.; Guillarme, D. Recent Advances and Current Challenges in Hydrophilic Interaction Chromatography for the Analysis of Therapeutic Oligonucleotides. TrAC Trends in Analytical Chemistry 2024, 176, 117758.
(11) Lanzillotti, M. B.; Brodbelt, J. S. Nucleo-SAFARI: Automated Identification of Fragment Ions in Top-Down MS/MS Spectra of Nucleic Acids. Anal Chem 2024, 43, 28.
(12) Ryan, J. P.; Slysz, G. W.; Rye, P.; Stow, S. M.; Dodds, J. N.; Sausen, J.; Baker, E. S. Increasing Oligonucleotide Sequencing Information and Throughput with Ion Mobility Spectrometry-Mass Spectrometry. J Am Soc Mass Spectrom 2025.
Acknowledgements: The author declares no conflicts of interest.
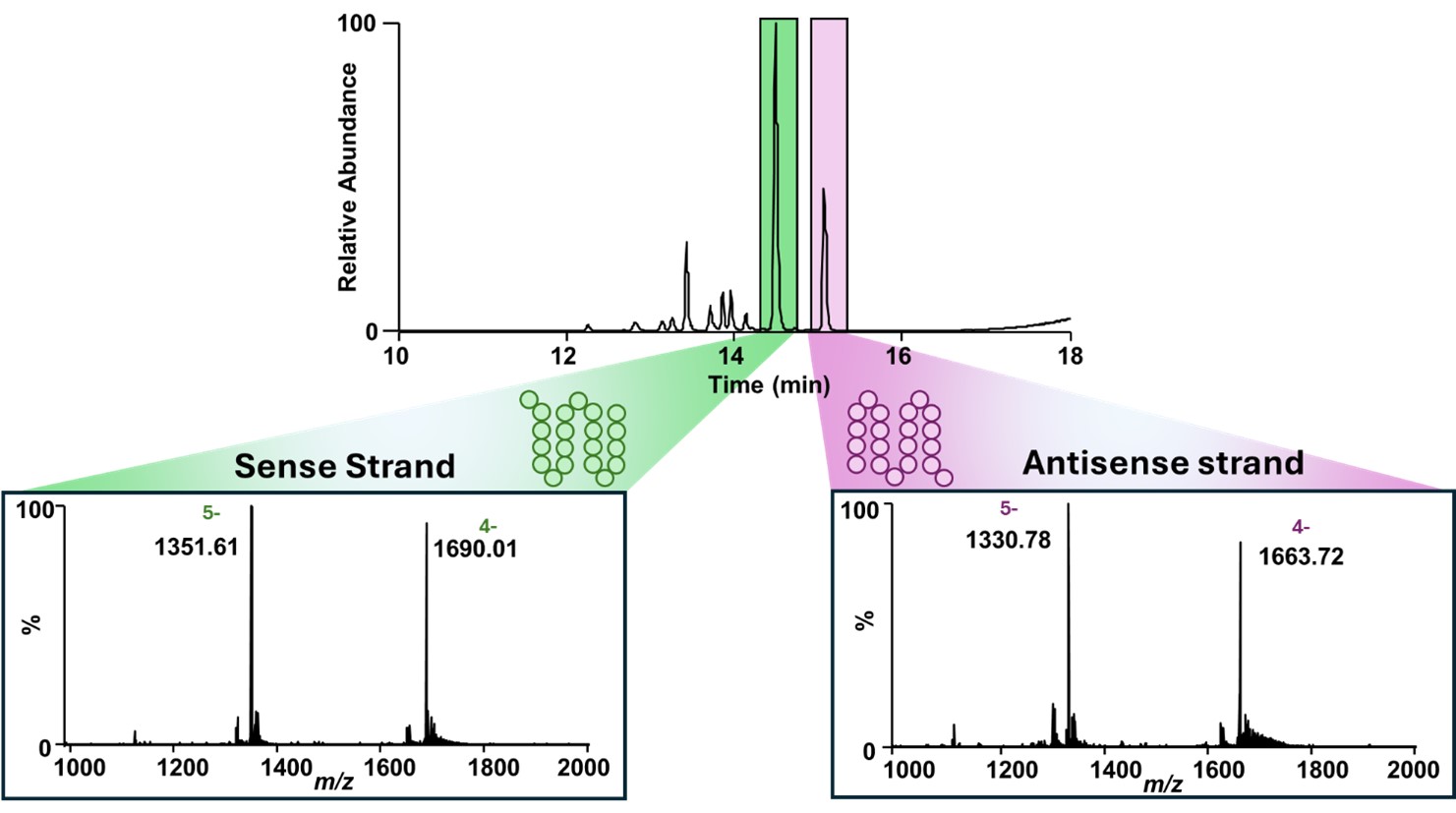 Figure 1. Separation and detection of sense and antisense strands of Patisiran siRNA using HILIC-MS. The GTx Resolve BEH Amide column enables baseline resolution of the sense (green) and antisense (purple) strands under MS-compatible conditions using 10 mM ammonium acetate. Inset graphs represent MS1 spectra acquired for each peak and confirm the identity of the individual strands based on their intact molecular mass.
Figure 1. Separation and detection of sense and antisense strands of Patisiran siRNA using HILIC-MS. The GTx Resolve BEH Amide column enables baseline resolution of the sense (green) and antisense (purple) strands under MS-compatible conditions using 10 mM ammonium acetate. Inset graphs represent MS1 spectra acquired for each peak and confirm the identity of the individual strands based on their intact molecular mass.
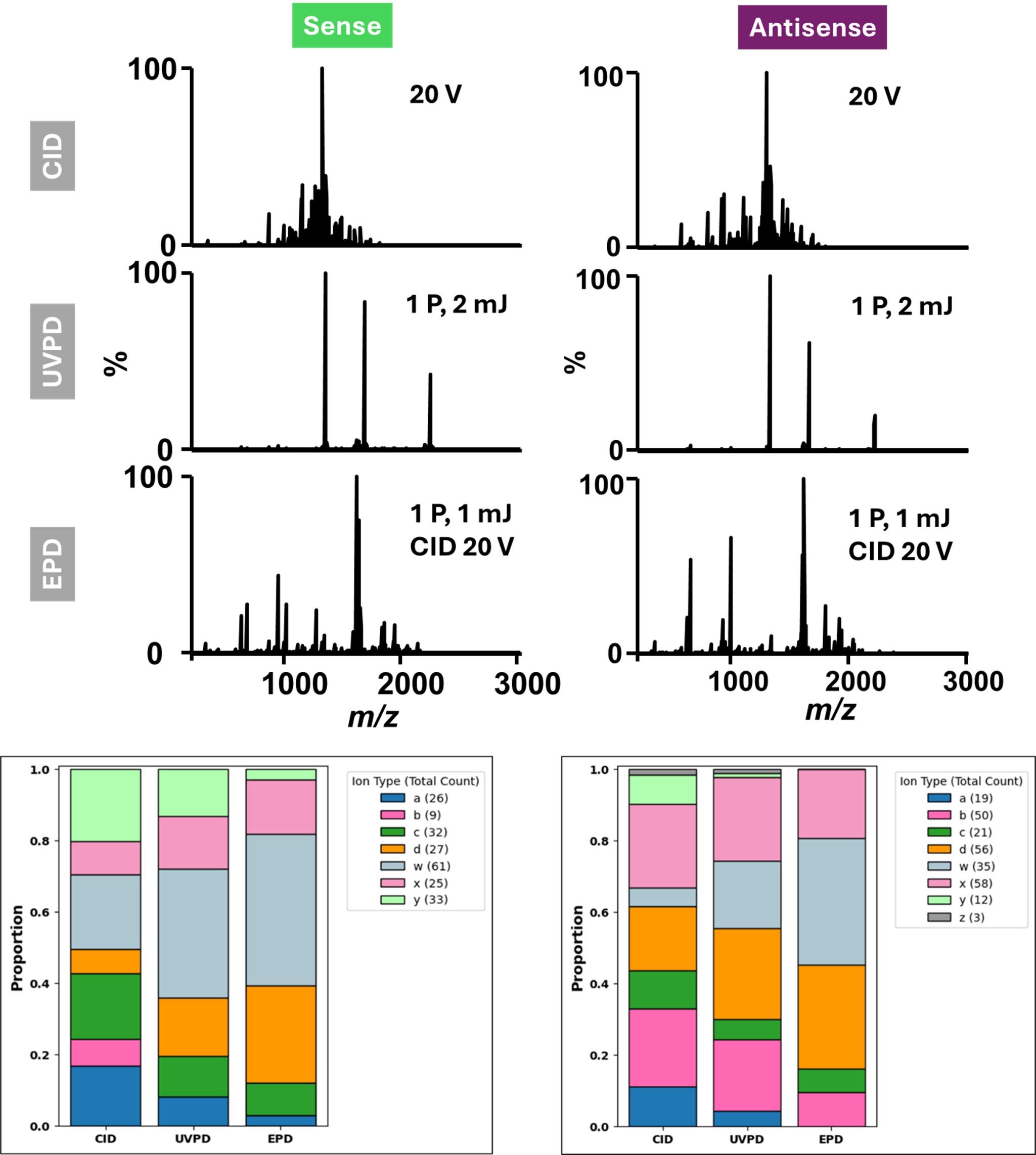 Figure 2. Comparison of gas-phase activation methods for structural characterization of sense and antisense strands of Patisiran siRNA. (A) Representative MS/MS spectra of the sense (left) and antisense (right) strands acquired using three different activation methods: CID (20 V), UVPD (1 laser pulse, 2 mJ), and EPD (1 laser pulse, 1 mJ + CID 20 V). (B) Ion-type distributions for each activation method show the relative contribution of different fragment series.
Figure 2. Comparison of gas-phase activation methods for structural characterization of sense and antisense strands of Patisiran siRNA. (A) Representative MS/MS spectra of the sense (left) and antisense (right) strands acquired using three different activation methods: CID (20 V), UVPD (1 laser pulse, 2 mJ), and EPD (1 laser pulse, 1 mJ + CID 20 V). (B) Ion-type distributions for each activation method show the relative contribution of different fragment series.
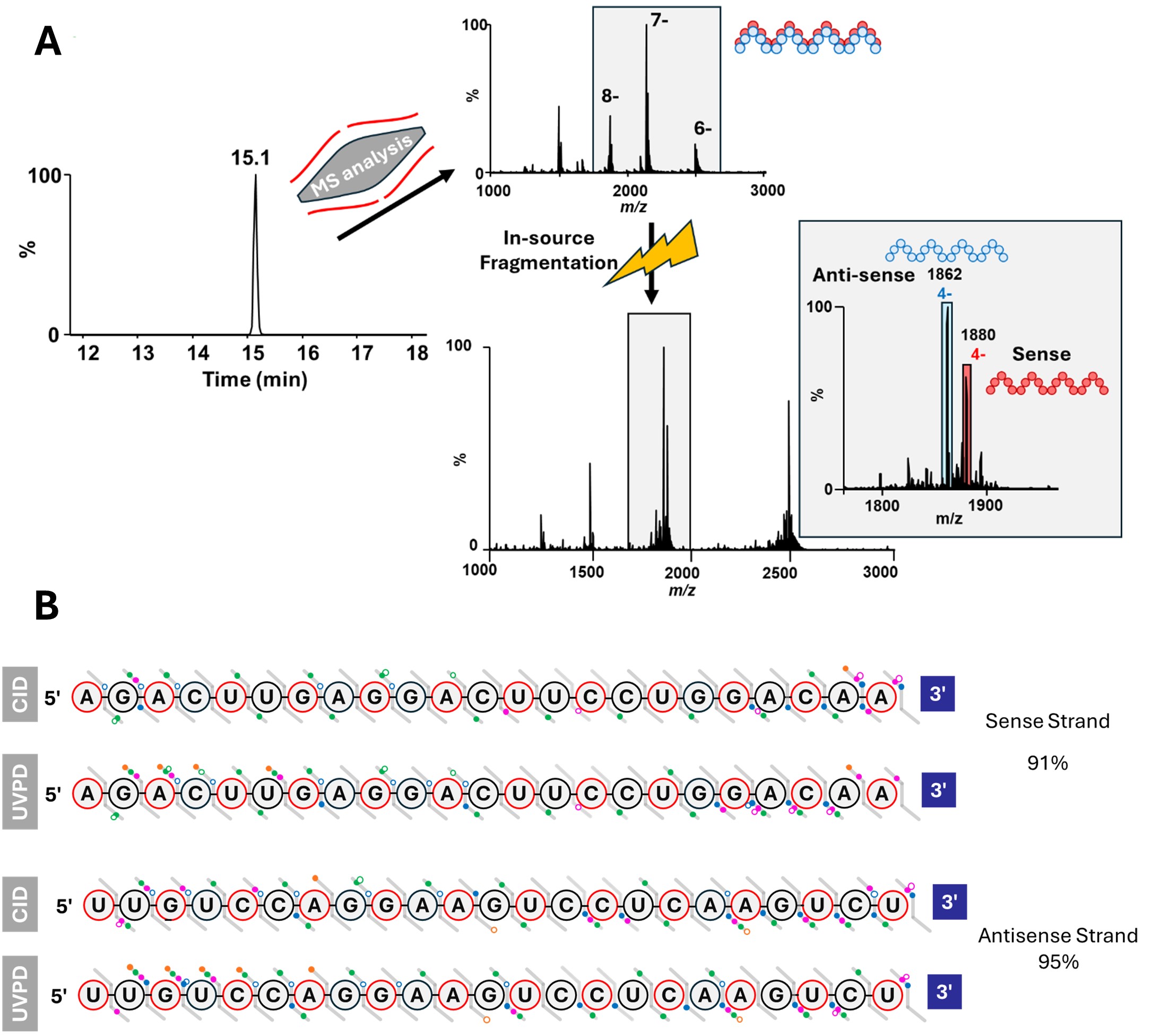 Figure 3. HILIC Separation of Duplex siRNA Enables Strand-Specific MS Analysis and High-Coverage Structural Characterization. (A) Extracted ion chromatogram showing a single peak at 15.1 min corresponding to intact duplex siRNA, preserved under native HILIC-MS conditions. In-source fragmentation was used to dissociate the duplex into its constituent sense and antisense strands followed by quadrupole isolation and identification of the individual RNA strands. (B) Combined CID and UVPD fragmentation maps show high sequence coverage of both strands.
Figure 3. HILIC Separation of Duplex siRNA Enables Strand-Specific MS Analysis and High-Coverage Structural Characterization. (A) Extracted ion chromatogram showing a single peak at 15.1 min corresponding to intact duplex siRNA, preserved under native HILIC-MS conditions. In-source fragmentation was used to dissociate the duplex into its constituent sense and antisense strands followed by quadrupole isolation and identification of the individual RNA strands. (B) Combined CID and UVPD fragmentation maps show high sequence coverage of both strands.
Methods: Chemically-modified Patisiran RNA strands were obtained from Integrated DNA Technologies (Coralville, IA), ATU027 and Fitusiran siRNAs were purchased from MedChemExpress. Samples were diluted to a final concentration of 10 µM in 10 mM piperidine and used without further purification. HILIC separation of therapeutic RNA was performed using a GTxResolve Premier BEH Amide column (300 Å, 1.7 µm, 2.1 × 150 mm; Waters) on a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific) using 25% acetonitrile (MPA) and 95% LC-MS acetonitrile (MPB) both with 10 mM ammonium acetate. MS analysis was performed using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) source operating in negative ion mode. The mass spectrometer was modified with a 193 nm excimer laser (Coherent Inc., 500 Hz) to enable ultraviolet photodissociation (UVPD) for MS/MS. For structural characterization, collision induced dissociation (CID) was performed using NCE ranging from 15 to 20. UVPD was conducted using a single laser pulse at 2-3 mJ to generate diverse fragment ions, while activated-electron photodetachment (a-EPD) was achieved with a single 1 mJ laser pulse followed by supplemental collisional activation (NCE 10–15). Mass spectral data were processed and interpreted using the Nucleo-SAFARI software platform.11
Results: A HILIC-MS workflow was established for the analysis of modified therapeutic RNAs, enabling effective separation of the sense and antisense strands of Patisiran without reliance on non-volatile ion-pairing reagents (Figure 1). The use of 10 mM ammonium acetate as a volatile mobile phase additive provided excellent chromatographic performance, yielding sharp, well-resolved peaks and minimizing salt adduction—thereby enhancing compatibility with mass spectrometric detection. CID of single strands achieved complete (100%) sequence coverage for both sense and antisense RNA, with fragmentation dominated by c- and y-type ions and a large amount of base-loss products (Figure 2). This indicates highly efficient backbone cleavage and confirms that CID is well suited for routine characterization of canonical phosphodiester backbones. In contrast, UVPD and EPD produced a more diverse array of fragment ions—including a-, w-, and d-type species as well as c- and y- fragments—which provided broader structural coverage across the RNA molecule. These techniques enabled the mapping of modifications located along the phosphate backbone, ribose sugar, and nitrogenous bases, offering a more comprehensive representation of RNA architecture than CID alone. When analyzing double-stranded siRNA, the HILIC method successfully preserved the native duplex, observed as a distinct peak in the chromatogram. Gentle in-source activation disrupted the duplex with minimum fragmentation, and quadrupole isolation using narrow m/z windows enabled strand-specific isolation prior to MS/MS (Figure 3). Under these conditions, CID provided 75% and 87% sequence coverage for the sense and antisense strands, respectively. However, the combination of CID and UVPD increased sequence coverage to 91% for the sense and 95% for the antisense strands—surpassing previously reported values for similar sizes of RNAs and highlighting the advantages of multimodal activation.12 Collectively, these results demonstrate that coupling HILIC-MS with complementary gas-phase activation techniques enables structural characterization of therapeutic RNA. This integrated approach offers a significant improvement over conventional IP-RPLC–MS workflows and is well suited for both discovery-stage and regulatory applications in RNA therapeutic development.
Conclusion: HILIC separation enabled effective resolution of intact therapeutic RNA strands from low-abundance degradation products without the need for non-volatile ion-pairing reagents, ensuring full compatibility with mass spectrometry detection. Coupling this chromatographic platform with advanced ion activation techniques provided comprehensive structural characterization, supporting accurate sequencing and validation of chemical modifications critical for RNA therapeutic development and quality assessment.
References: (1) Androsavich, J. R. Frameworks for Transformational Breakthroughs in RNA-Based Medicines. Nature Reviews Drug Discovery 2024 23:6 2024, 23 (6), 421–444.
(2) Saw, P. E.; Song, E. Advancements in Clinical RNA Therapeutics: Present Developments and Prospective Outlooks. Cell Rep Med 2024, 5 (5), 101555.
(3) Shi, Y.; Zhen, X.; Zhang, Y.; Li, Y.; Koo, S.; Saiding, Q.; Kong, N.; Liu, G.; Chen, W.; Tao, W. Chemically Modified Platforms for Better RNA Therapeutics. Chem Rev 2024, 124 (3), 929–1033.
(4) Delaunay, S.; Helm, M.; Frye, M. RNA Modifications in Physiology and Disease: Towards Clinical Applications. Nature Reviews Genetics 2023 25:2 2023, 25 (2), 104–122.
(5) Li, Q.; Yin, K.; Ma, H. P.; Liu, H. H.; Li, S.; Luo, X.; Hu, R.; Zhang, W. W.; Lv, Z. S.; Niu, X. L.; Gu, M. H.; Li, C. L.; Liu, Y. S.; Liu, Y. J.; Li, H. B.; Li, N.; Li, C.; Gu, W. W.; Li, J. J. Application of Improved GalNAc Conjugation in Development of Cost-Effective SiRNA Therapies Targeting Cardiovascular Diseases. Molecular Therapy 2024, 32 (3), 637–645.
(6) Kashyap, D.; Booth, M. J. Nucleic Acid Conjugates: Unlocking Therapeutic Potential. ACS Bio and Med Chem Au 2024.
(7) Maurer, J.; Malburet, C.; François-Heude, M.; Guillarme, D. Evaluation of Ion Pairing Reversed-Phase Liquid Chromatography for the Separation of Large RNA Molecules. J Chromatogr A 2025, 1740, 465574.
(8) Hayashi, Y.; Sun, Y. Overcoming Challenges in Oligonucleotide Therapeutics Analysis: A Novel Nonion Pair Approach. J Am Soc Mass Spectrom 2024, 35 (9), 2034–2037.
(9) Hsiao, J. J.; Bertram, L. J.; Apffel, A.; Tripodi, A. A. P.; Coffey, A.; Wei, T.-C.; Flannery, C. Systematic Evaluation of HILIC Stationary Phases for MS Characterization of Oligonucleotides. LCGC International 2024, 10–20.
(10) Lardeux, H.; D’Atri, V.; Guillarme, D. Recent Advances and Current Challenges in Hydrophilic Interaction Chromatography for the Analysis of Therapeutic Oligonucleotides. TrAC Trends in Analytical Chemistry 2024, 176, 117758.
(11) Lanzillotti, M. B.; Brodbelt, J. S. Nucleo-SAFARI: Automated Identification of Fragment Ions in Top-Down MS/MS Spectra of Nucleic Acids. Anal Chem 2024, 43, 28.
(12) Ryan, J. P.; Slysz, G. W.; Rye, P.; Stow, S. M.; Dodds, J. N.; Sausen, J.; Baker, E. S. Increasing Oligonucleotide Sequencing Information and Throughput with Ion Mobility Spectrometry-Mass Spectrometry. J Am Soc Mass Spectrom 2025.
Acknowledgements: The author declares no conflicts of interest.
 Figure 1. Separation and detection of sense and antisense strands of Patisiran siRNA using HILIC-MS. The GTx Resolve BEH Amide column enables baseline resolution of the sense (green) and antisense (purple) strands under MS-compatible conditions using 10 mM ammonium acetate. Inset graphs represent MS1 spectra acquired for each peak and confirm the identity of the individual strands based on their intact molecular mass.
Figure 1. Separation and detection of sense and antisense strands of Patisiran siRNA using HILIC-MS. The GTx Resolve BEH Amide column enables baseline resolution of the sense (green) and antisense (purple) strands under MS-compatible conditions using 10 mM ammonium acetate. Inset graphs represent MS1 spectra acquired for each peak and confirm the identity of the individual strands based on their intact molecular mass. Figure 2. Comparison of gas-phase activation methods for structural characterization of sense and antisense strands of Patisiran siRNA. (A) Representative MS/MS spectra of the sense (left) and antisense (right) strands acquired using three different activation methods: CID (20 V), UVPD (1 laser pulse, 2 mJ), and EPD (1 laser pulse, 1 mJ + CID 20 V). (B) Ion-type distributions for each activation method show the relative contribution of different fragment series.
Figure 2. Comparison of gas-phase activation methods for structural characterization of sense and antisense strands of Patisiran siRNA. (A) Representative MS/MS spectra of the sense (left) and antisense (right) strands acquired using three different activation methods: CID (20 V), UVPD (1 laser pulse, 2 mJ), and EPD (1 laser pulse, 1 mJ + CID 20 V). (B) Ion-type distributions for each activation method show the relative contribution of different fragment series.  Figure 3. HILIC Separation of Duplex siRNA Enables Strand-Specific MS Analysis and High-Coverage Structural Characterization. (A) Extracted ion chromatogram showing a single peak at 15.1 min corresponding to intact duplex siRNA, preserved under native HILIC-MS conditions. In-source fragmentation was used to dissociate the duplex into its constituent sense and antisense strands followed by quadrupole isolation and identification of the individual RNA strands. (B) Combined CID and UVPD fragmentation maps show high sequence coverage of both strands.
Figure 3. HILIC Separation of Duplex siRNA Enables Strand-Specific MS Analysis and High-Coverage Structural Characterization. (A) Extracted ion chromatogram showing a single peak at 15.1 min corresponding to intact duplex siRNA, preserved under native HILIC-MS conditions. In-source fragmentation was used to dissociate the duplex into its constituent sense and antisense strands followed by quadrupole isolation and identification of the individual RNA strands. (B) Combined CID and UVPD fragmentation maps show high sequence coverage of both strands. 